Metagenomi Technologies IPO investment analysis
January 8, 2024
This is not investment advice. We used AI and automated software tools for most of this research. A human formatted the charts based on data / analysis from the software, prompted the AI to do some editing, and did some light manual editing. We did some fact checking but cannot guarantee the accuracy of everything in the article. We do not have a position in or an ongoing business relationship with the company.
Overview
Metagenomi is a company specializing in genetic medicine, focusing on developing treatments for genetic diseases using a unique genome editing toolbox derived from metagenomics. This approach involves analyzing genetic material from various natural environments to discover and utilize microbial genetic diversity.
The platform includes an array of tools such as programmable nucleases (including type II and type V Cas nucleases and SMall Arginine-Rich sysTems or SMART nucleases), base editors, RNA and DNA-mediated integration systems, prime editing systems, and CRISPR-associated transposases (CASTs). The platform leverages environmental DNA from diverse ecosystems, which is deeply sequenced to identify novel proteins and CRISPR systems. This has led to the discovery of over 20,000 new genome editing systems. The company employs high-throughput screening, artificial intelligence, and proprietary algorithms to mine through billions of novel proteins for creating genome editing tools.
Metagenomi's tools are designed for site-specific genome editing. This includes programmable nucleases for precise DNA cutting, base editors for single nucleotide changes, and systems for both small and large genomic integrations. The tools have been extensively evaluated in various models, including human primary cells and nonhuman primates, demonstrating efficacy and addressing limitations like off-target effects. The platform's tools are compatible with a range of delivery methods, both viral and nonviral, due to the varied sizes and biochemistry of the nucleases and guide RNAs.
Metagenomi filed to go public in January 2024. This note includes an analysis of the potential IPO valuation.
Pipeline overview
| Product name | Modality | Target | Indication | Discovery | Preclinical | Phase 1 | Phase 2 | Phase 3 | FDA submission | Commercial |
|---|---|---|---|---|---|---|---|---|---|---|
| Hemophilia A program | Gene editing | FVIII knock in | Hemophilia A | |||||||
| Primary Hyperoxaluria Type 1 program | Gene editing | HAO1 knock down | Primary Hyperoxaluria Type 1 | |||||||
| Transthyretin Amyloidosis program | Gene editing | TTR knock down | Transthyretin Amyloidosis | |||||||
| Cardiovascular Disease program | Gene editing | AGT knock down | Cardiovascular Disease | |||||||
| Alpha 1 Antitrypsin Deficiency program | Gene editing | A1AT RIGS/Base editing | Alpha 1 Antitrypsin Deficiency | |||||||
| Wilson's Disease program | Gene editing | ATP7B RIGS | Wilson's Disease | |||||||
| Familial ALS program | Gene editing | SOD1 Knock down | Familial ALS | |||||||
| Spontaneous ALS program | Gene editing | ATXN2 Knock down | Spontaneous ALS | |||||||
| Charcot-Marie-Tooth Type 1A program | Gene editing | PMP22 Knock down | Charcot-Marie-Tooth Type 1A | |||||||
| Duchenne Muscular Dystrophy program | Gene editing | DMD Exon skipping/CAST | Duchenne Muscular Dystrophy | |||||||
| Cystic Fibrosis program | Gene editing | CFTR CAST | Cystic Fibrosis | |||||||
| Immuno-oncology cell therapy programs | Cell therapy | Multiple | Oncology | |||||||
| Autoimmune CAR-T program | Cell therapy | Multiple | Autoimmune disease |
Highlights and risks
Broad platform of gene editors including base editors, prime editors and systems for large-scale integration
Potential best-in-class editors, including some of the smallest base editors to date
Platform of over 20,000 new genome editing systems provides potential for differentiated, unique systems
Large pipeline of a variety of gene editing programs targeting validated indications
Early-stage pipeline with no programs in clinical trials
Targeting validated, but competitive indications creates a high bar for competitive differentiation
Development of broad pipeline will require significant capital and partnerships, and prioritizing asset development will be critical
Valuation
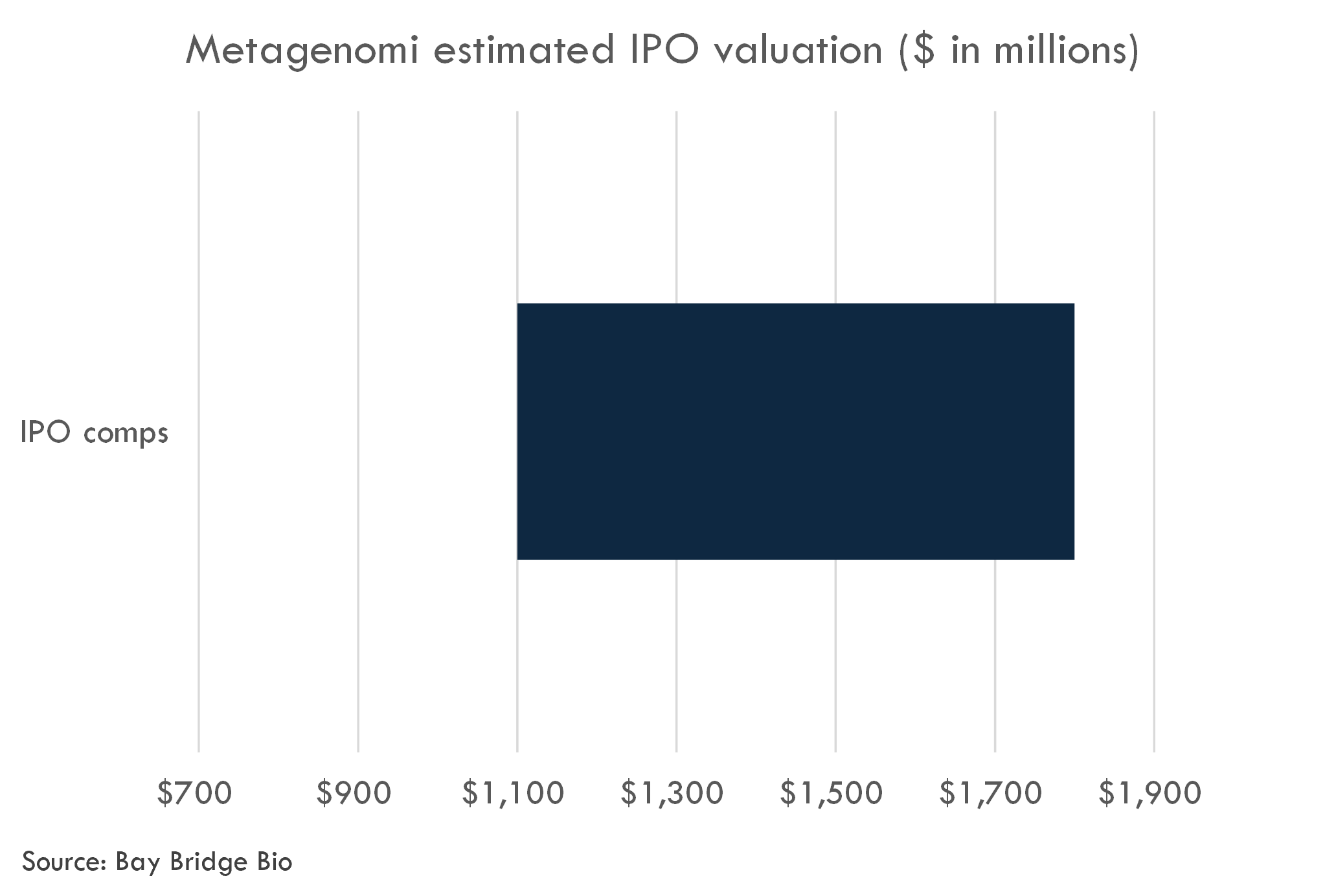
Metagenomi filed to go public in January 2024. The company has riased over $350 million to date. We estimate the post-money valuation of the last Series B-1 round at $806 million.
Based on comparable IPOs, we estimate an IPO valuation range of $1.1 billion to $1.8 billion, on a fully diluted post-money basis. We estimate that the company could raise $200-300 million in the IPO.
Due to the early-stage of the pipeline, we did not conduct a DCF analysis. We anticipate that the company could potentially become an M&A target after demonstrating clinical proof-of-concept, but due to the early-stage nature of the gene editing field, there have not been any large M&A transactions.
Based on recent IPOs, we estimate the following investors could be interested in Metagenomi's IPO:

Weekly analyses of biotech startups, generated by AI
Receive high quality, AI-generated analyses of biotech startups, public companies and scientific papers each week.
Major Translational Programs
Hemophilia A Program
This program focuses on a novel, durable, knock-in method to enable the expression of the clotting Factor VIII (FVIII). Instead of providing the FVIII gene episomally, which could be unstable, Metagenomi's approach is to insert a FVIII DNA cassette into a 'safe harbor location' within the albumin gene. This allows for the expression of FVIII driven by the strong albumin promoter.
Feasibility has been shown in mice, and ongoing studies in nonhuman primates (NHP) have shown promising results for up to 4.5 months post-treatment. The goal is to establish long-term FVIII expression.
Next steps include comparison of different FVIII donor DNA cassettes in NHP studies to select a development candidate by Q2 2024, and preparation for IND-enabling studies.
Primary Hyperoxaluria, Type 1 (PH1) Program
The program aims for a durable knockdown of HAO1 gene expression to reduce the substrate in PH1, a metabolic disorder leading to kidney failure. Optimal nuclease and gRNA combinations have been screened, and preclinical proof-of-concept has been achieved in a mouse model.
Next steps include completion of candidate confirmation for NHP studies, with NHP data expected in 2024 to support final development candidate selection.
Transthyretin Amyloidosis Program
The goal is to knock down TTR gene expression in patients with this disease, where misfolded proteins aggregate and cause organ dysfunction. More than 90% knockdown of human TTR protein has been achieved after a single dose in a humanized TTR mouse model.
Next steps include advanced nuclease and guide selection, with plans to move into NHP studies in 2024.
Further Areas of Focus
- Liver-Associated Targets: Efforts are being made to deliver novel RIGS to the liver to potentially correct mutations in Wilson’s disease and alpha-1-antitrypsin deficiency (A1AT deficiency). A base editor approach is being explored for A1AT deficiency due to its single base pair predominant mutation.
- Neurodegenerative and Neuromuscular Diseases: Expansion of the technology to address central nervous system and muscle targets, with the initial focus on neurodegenerative and neuromuscular diseases. This includes amyotrophic lateral sclerosis (SOD1, ATXN2) and Charcot-Marie-Tooth Type 1a (PMP22), as well as exon skipping approaches for Duchenne Muscular Dystrophy.
- Cystic Fibrosis: Developing larger RNA- and DNA-mediated integration systems to potentially provide a single curative approach for cystic fibrosis by delivering a full copy of a functional CFTR gene.
These programs are all in the preclinical stage, with no approved products yet. The goal of these programs is to leverage Metagenomi's genome editing toolbox to develop therapies that could offer long-term solutions to these diseases, supported by validating preclinical and clinical data, with a focus on meeting significant clinical needs.
Platform overview
Metagenomi's platform includes a variety of gene editing components:
- Programmable Nucleases (MG Type II & V and SMART Nucleases): These are versatile editing tools guided by RNA to target specific genome locations for cutting. They are the foundation for genome editing, enabling knock-ins and knockdowns.
- Base Editors (MG ABE and MG CBE): These are engineered to perform nucleotide changes, including adenine and cytosine deaminase-mediated edits, for precise single base pair alterations.
- RNA-mediated Integration Systems (MG Little RIGS and MG Big RIGS): These systems facilitate small corrections (Little RIGS) and large genomic integrations (Big RIGS) without needing double-stranded DNA breaks.
- CRISPR-associated Transposases (MG CAST): They offer DNA-mediated integration capabilities for potentially large templates, expanding the scope of what can be achieved compared to RIGS.
Their approach offers several advantages:
- Efficiency and Precision: The toolbox provides efficient and precise genome editing capabilities, with diverse nucleases for extensive genome targeting.
- Modularity: The tools are modular and can be matched to disease-specific targets, with the ability to edit any codon in the human genome.
- Compatibility: Designed to be compatible with both viral and nonviral delivery technologies, including small guides for LNP approaches and systems that fit within the constraints of AAV vectors.
- Preclinical Validation: The nucleases have shown broad potency in various models, including human, mouse, and nonhuman primates, with minimal off-target effects.
- Diverse Editing Capabilities: The platform can address a wide variety of genomic modifications, from single nucleotide changes to large genomic integrations.
- Potential for Universal Treatment: The integration systems could potentially allow for a single genetic medicine to treat all patients with a disease, particularly those caused by loss-of-function mutations.
Metagenomi's platform represents a comprehensive and advanced system for genome editing, with the potential to address complex genetic diseases through a variety of sophisticated and precise tools.
Overview of Gene Editing
Gene editing is a revolutionary technology that enables precise, directed changes to genomic DNA. At its core, it involves the use of engineered nucleases, or "molecular scissors," to make targeted cuts in the DNA strand. This allows for the deletion, insertion, or alteration of specific DNA sequences. Key technologies in this field include CRISPR-Cas9, TALENs (Transcription Activator-Like Effector Nucleases), and ZFNs (Zinc Finger Nucleases). CRISPR-Cas9, in particular, has garnered widespread attention due to its ease of use, efficiency, and versatility.
Some notable gene editing products and their applications include CRISPR Therapeutics' CTX001 for the treatment of beta thalassemia and sickle cell disease, and Editas Medicine's EDIT-101 for Leber Congenital Amaurosis, a rare genetic eye disease. These products represent the forefront of gene editing technologies being applied in clinical settings.
Compared to traditional therapeutic approaches, gene editing offers a high degree of specificity and the potential for a one-time curative treatment. This approach can directly target the root cause of genetic diseases, rather than just treating symptoms. It's particularly advantageous in diseases caused by a single gene defect. Gene editing also reduces the long-term costs and complications associated with ongoing treatments like enzyme replacement therapies.
The potential of gene editing extends beyond monogenic diseases. It holds promise for the treatment of more complex conditions like cancer, HIV, and neurodegenerative diseases. In oncology, gene editing could be used to engineer immune cells to better target and destroy cancer cells. The technology is also being explored in the development of antiviral strategies, particularly for diseases like HIV, where the virus integrates into the host genome. Furthermore, ongoing research is investigating the use of gene editing for regenerative medicine, such as repairing damaged tissues and organs.
While the full potential of gene editing is yet to be realized, its capability to provide precise, personalized, and potentially permanent treatments for a range of diseases marks it as a transformative advance in the field of medicine.
Limitations of Gene Editing Technologies
Various challenges are faced by traditional CRISPR-based approaches, as well as other methods like NHEJ, HDR, base editing, prime editing, serine integrases, PASTE technology, and more. Here's a summary:
- Complexity of Editing: Many current platforms are specialized for specific types of edits, such as gene knockouts, single-nucleotide edits, or small gene corrections. This specialization limits the range of diseases they can address and restricts their ability to perform diverse types of genome modifications.
- Specificity and Control: There is a need for high specificity to avoid off-target effects, which could lead to unintended mutations and potential safety concerns. Achieving precise genome editing without affecting other parts of the genome remains a significant challenge.
- Delivery Size Constraints: The size of the most widely used SpCas9 nuclease is large, about 1,300 amino acids, making it difficult to package into some delivery vectors like AAV, which are used to target tissues other than the liver.
- Target Sequence Limitations: SpCas9 requires a specific 'NGG' protospacer adjacent motif (PAM) sequence, limiting its targeting range within the genome and, by extension, the range of mutations it can correct.
- Engineering Requirements: To achieve therapeutically relevant activity, extensive engineering of naturally occurring nucleases and effectors is often necessary, which can lead to significant delays in the development process.
- Licensing Constraints: Many gene editing technologies have been developed under specific licensing agreements from academic institutions, limiting their use to the terms of those agreements.
- Immune Responses: The immune system may recognize and react against the bacterial-derived Cas proteins or the delivery vectors, potentially leading to inflammatory responses or reduced editing efficiency.
- In Vivo Editing Challenges: Achieving high editing efficiency in vivo remains a hurdle, especially in tissues that are not easily transfected or transduced, such as the brain or bone marrow.
- Prolonged Expression and Re-administration: For viral delivery systems like AAV, the prolonged expression of nuclease may lead to sustained DNA damage, and re-administration is limited due to immune responses against the viral capsid.
- Mosaicism: Incomplete or uneven editing across a population of cells can lead to mosaicism, where both edited and unedited cells coexist, potentially reducing the therapeutic benefit.
- Scalability and Manufacturability: Scaling up production for clinical applications while maintaining quality and activity of the gene editing components can be challenging.
- Cost: The high cost of developing, manufacturing, and delivering gene editing therapies may limit their accessibility and adoption.
Beyond these general limitations, specific gene editing techniques have unique challenges:
- Traditional CRISPR-Cas Systems: Often lead to random insertions or deletions (indels) during the DNA repair process, which may not be suitable for precise gene correction therapies.
- NHEJ (Non-homologous End Joining): An error-prone repair mechanism that can lead to indels at the cut site, potentially causing unwanted mutations.
- HDR (Homology-Directed Repair): More precise than NHEJ but has low efficiency in most cell types and is challenging in non-dividing cells.
- Base Editing: Induces single base pair conversions without double-stranded DNA breaks but is limited to certain types of point mutations and can have off-target deamination activity.
- Prime Editing: Allows for a wider variety of DNA edits without double-stranded breaks but is complex and requires further development for efficiency and reduced off-target activity.
- Serine Integrases / PASTE Technology: Allows for DNA sequence integration but typically has restricted targeting capabilities and requires significant protein engineering.
Metagenomi's technology can potentially address the previously mentioned limitations:
- Complexity of Editing
Metagenomi's diverse suite of tools could potentially allow for a range of genome modifications beyond simple knockouts and single-nucleotide edits, addressing the complexity of editing. However, the successful application of these tools to complex genomic rearrangements in a therapeutic context remains to be proven.
- Specificity and Control
The specificity of their tools, if as high as suggested, could reduce off-target effects. However, it's important to note that even with highly specific nucleases, off-target effects can never be entirely ruled out and must be carefully evaluated in clinical settings.
- Delivery Size Constraints
By discovering enzymes of various sizes, Metagenomi may overcome some of the delivery constraints posed by larger nucleases like SpCas9. Still, the efficiency of delivery to certain tissues and crossing biological barriers like the blood-brain barrier could pose challenges.
- Target Sequence Limitations
If Metagenomi's nucleases can indeed target a wider range of sequences without the strict PAM requirements of SpCas9, this could significantly expand the targetable genome. However, this potential needs to be validated experimentally and in clinical trials.
- Engineering Requirements
The platform's ability to identify naturally occurring enzymes that require minimal engineering is promising. However, some degree of customization will likely always be needed to tailor these systems to specific therapeutic needs.
- Licensing Constraints
Metagenomi's proprietary platform could provide freedom to operate independently of the complex CRISPR patent landscape. However, as they progress, they may still face challenges related to intellectual property, including potential patent disputes.
- Efficiency of HDR
Metagenomi's technology does not directly address the inherent efficiency issues of HDR, which is a limitation of the cell's natural repair mechanisms rather than the genome editing tools themselves.
- Immune Responses
The immune response to foreign proteins is a biological response that could affect any gene editing tool. Metagenomi's technology might still face immune challenges, especially if novel nucleases are immunogenic.
- In Vivo Editing Challenges
Metagenomi's approach could potentially allow for effective in vivo editing if their enzymes are suitably efficient and can be delivered effectively. However, the in vivo delivery of these tools to certain tissues remains a significant hurdle for all gene editing technologies.
- Prolonged Expression and Re-administration
Issues with prolonged expression and limitations on re-administration are related to the delivery vector and immune response, which may not be fully resolved by Metagenomi's nucleases.
- Mosaicism
The potential for mosaicism is a challenge for all gene editing technologies, and Metagenomi's approach does not inherently solve this issue. It will depend on the efficiency and cell-type specificity of their tools.
- Ethical and Regulatory Considerations
These concerns are applicable to all gene editing endeavors and are not specifically addressed by Metagenomi's technology.
- Scalability and Manufacturability
Metagenomi's platform may streamline the discovery and development process, but scaling up production for clinical use is a challenge that remains to be addressed.
- Cost Metagenomi's methods may reduce some costs associated with engineering and development, but gene editing therapies are inherently expensive, and it's uncertain how much cost reduction can be achieved.
In summary, Metagenomi's platform presents a promising approach to addressing several limitations of current gene editing technologies, especially in terms of diversity and specificity of tools, and targeting flexibility. However, there are inherent challenges in the field of gene therapy, such as delivery, immune response, and HDR efficiency, that are not fully overcome by any current technology, including Metagenomi's. Continued research and clinical trials will be crucial to assess the efficacy and safety of their platform.
Platform in depth
Programmable nucleases
Metagenomi's approach to identifying programmable nucleases is multifaceted and innovative, leveraging the diversity of microbial life to find novel genome editing tools. Here's how they do it:
Metagenomi uses a proprietary database built from environmental samples to discover a vast array of novel CRISPR systems. This database is expected to contain novel sequences that have evolved in various microorganisms, providing a rich source of potential new genome editing enzymes.
Their attention is on type II and type V CRISPR systems due to their simplicity and programmability. These systems use RNA-guided nucleases that can target specific DNA sequences and, in some cases, RNA sequences. They have found several new types of ultra-small nucleases, which they refer to as SMART, along with novel type V sub-groups. These are considerably smaller than the commonly used SpCas9, making them potentially more suitable for certain delivery vectors.
To validate the activity of these newly discovered nucleases, Metagenomi performs high-throughput in vitro testing. This process involves bioinformatic predictions followed by experimental validation in cell-free systems and mammalian cells. Their screening process emphasizes finding natural, un-engineered nucleases that show high activity and specificity. They aim to identify systems that perform as well or better than SpCas9 with fewer off-target effects. They evaluate the targeting density of their nucleases, which refers to the frequency of targetable sites within the human genome. A higher targeting density increases the chances of finding an effective nuclease-guide RNA combination for any given genomic target.
Beyond simple genome editing, the programmable nucleases they discover can serve as platforms for developing more specialized tools like base editors and RNA-mediated integration systems (RIGS). They also use a modular approach to create chimeric systems that combine the high activity of top-performing nucleases with the unique targeting domains from other systems. This allows them to tailor their tools to specific genomic targets.
Metagenomi's technology has the potential to overcome some limitations of current gene editing technologies. Their smaller, potentially more specific nucleases could address delivery size constraints and target sequence limitations. The high-throughput screening process and focus on specificity could help minimize off-target effects. However, challenges such as in vivo delivery, immune responses, and the efficiency of homology-directed repair (HDR) are not specifically addressed and remain areas where Metagenomi's approach will need to demonstrate efficacy. Additionally, the translation of these discoveries into clinical applications is a complex process that will require further development and validation.
Metagenomi's approach to gene editing using a diverse array of programmable nucleases from their metagenomics platform offers several potential advantages over established nucleotide editing approaches:
- Broader Targeting Range: By leveraging CRISPR type II and type V systems, Metagenomi's nucleases could potentially target a wider range of genomic sequences compared to established nucleases that have more restrictive PAM requirements or target sequence limitations.
- Reduced Size for Easier Delivery: The identification of ultra-small nucleases could enable packaging within viral vectors like AAVs that have size constraints. This can facilitate the delivery of gene editing tools to a variety of tissues and cell types that are otherwise inaccessible with larger enzymes like SpCas9.
- High Specificity and Reduced Off-Target Effects: Their selection process emphasizes high specificity, which is crucial for minimizing off-target mutations—a significant concern with established CRISPR systems. This could lead to safer gene editing interventions.
- Natural Enzyme Efficiency: The use of natural, un-engineered nucleases that have evolved to be highly effective within their microbial environments may translate to more efficient gene editing in human cells without the need for extensive protein engineering.
- Versatility and Modularity: Metagenomi's modular engineering approach allows for the creation of chimeric nucleases with tailored properties. This versatility enables the design of nucleases for a wide array of therapeutic applications.
- Comprehensive Genome Coverage: With a high targeting density, Metagenomi’s nucleases could potentially address any genetic locus, opening the door to therapeutic interventions for a broader range of genetic diseases.
- Rapid Development Pipeline: Utilizing predictive algorithms and high-throughput screening, the development of new nucleases from identification to application may be faster than traditional methods, which often involve extensive trial and error.
- Independence from Licensing Constraints: Developing a unique suite of proprietary nucleases may offer Metagenomi more freedom to operate and could reduce dependency on the existing, complex CRISPR patent landscape.
These advantages suggest that Metagenomi's approach could complement and potentially surpass the capabilities of existing gene editing technologies in terms of precision, versatility, and applicability. However, these benefits are theoretical until proven in clinical settings, where real-world challenges such as delivery efficiency, immune responses, and long-term safety need to be thoroughly evaluated.
Weekly analyses of biotech startups, generated by AI
Receive high quality, AI-generated analyses of biotech startups, public companies and scientific papers each week.
The evidence presented by Metagenomi indicates that their nucleases have the potential to address several limitations of current gene editing technologies. However, while the data appears robust and promising, the translation from preclinical models to successful clinical applications will be the ultimate test of these advantages. It will be important for Metagenomi to demonstrate that the nucleases work effectively in humans, with sustained efficacy and without adverse effects over the long term. Additionally, while the in vitro and in vivo data are encouraging, issues such as delivery to specific tissues, immune responses, and long-term durability of the edits in the context of a living organism are complex challenges that still need to be navigated as development progresses.
Potential advantages of Metagenomi's Nucleases
Nucleases for gene editing face several limitations, some of which are inherent to the nature of the technology and some due to biological complexity. Here is an overview of these challenges and an assessment of how well Metagenomi's nucleases might overcome them:
Delivery to Specific Tissues Effective delivery to target tissues besides the liver, such as the brain or muscle, remains a challenge. Metagenomi's discovery of ultra-small nucleases could facilitate the use of delivery vectors with broader tissue tropism, potentially addressing this limitation.
Immunogenicity The human immune system can recognize and mount a response against the bacterial-derived Cas proteins used in gene editing, which may limit their effectiveness. Metagenomi's approach to identifying a diverse array of nucleases may uncover enzymes that are less immunogenic or amenable to modifications that reduce immunogenicity.
Durability of Editing Ensuring that gene edits are stable over the long term and do not revert or cause late-onset off-target effects is important. The specificity and high efficiency of Metagenomi's nucleases suggest they could create durable edits, but this needs to be confirmed over time in living organisms.
Efficiency in Different Cell Types Some nucleases work well in certain cell types but not others. Metagenomi's nucleases have shown high activity across a range of primary human cells, which is promising for their versatility.
Complex and Large-Scale Genomic Edits Achieving large-scale genomic insertions or complex rearrangements is difficult with current technology. While Metagenomi's platform shows promise for a variety of edits, the ability to perform large or complex rearrangements has not been specifically addressed.
In Vivo Re-administration The potential for immune responses against viral vectors can limit the ability to re-administer gene-editing components when necessary. Metagenomi's platform does not directly address this challenge, as it is more related to the delivery system than to the nuclease itself.
Ethical and Regulatory Hurdles Gene editing technology raises significant ethical and regulatory considerations, especially concerning germline editing. These are policy and public perception issues that are outside the technical scope of Metagenomi's platform.
Cost-Effectiveness The development, manufacturing, and delivery of gene-editing therapies can be expensive. While the use of Metagenomi's high-throughput screening and AI algorithms could streamline some aspects of the development process, it remains to be seen how this will impact the overall cost of therapies developed using their nucleases.
Scalability of Manufacturing Producing gene-editing components at a scale sufficient for widespread clinical use while maintaining quality is challenging. Metagenomi's methods may simplify the discovery process, but manufacturing scalability is a separate issue that is not directly addressed by their technology.
Cas9-Induced P53 Response There's evidence that the DNA damage response from Cas9 editing can activate the p53 pathway, which may select for p53-deficient cells, potentially leading to oncogenic outcomes. The specificity of Metagenomi's nucleases may reduce DNA damage and hence p53 activation, but this would need specific investigation.
In conclusion, Metagenomi's nucleases show potential to overcome several but not all of the limitations faced by current gene editing tools. Some challenges, particularly those related to delivery, immune response, and long-term safety and efficacy in humans, are areas where additional research and clinical data will be necessary to fully assess the capabilities and limitations of their technology.
Metagenomi's Base Editors
Metagenomi's approach to identifying base editors is rooted in the integration of their proprietary programmable nucleases with their vast metagenomics discovery platform. Here's how they are developing these base editors and how the evidence supports the potential advantages of their technology:
Metagenomi has created both Adenine Base Editors (ABEs) and Cytosine Base Editors (CBEs), which have been validated in mammalian cells and in vivo. These base editors consist of a targeting component (a modified nuclease) and a deaminase that chemically alters the specific bases. ABEs and CBEs offer the advantage of precise editing without inducing double-stranded breaks, a significant advancement over traditional CRISPR/Cas9 editing which often relies on creating such breaks.
The base editors are designed using modified nickases, programmed to make a single-strand cut in the DNA, allowing the deaminase to chemically modify the targeted bases. This avoids the potential complications associated with double-stranded DNA breaks, such as unwanted insertions or deletions.
Evidence of high editing efficiency is presented, showing that their ABEs and CBEs can achieve significant levels of editing. This includes up to 60% editing efficiency in primary hepatocytes and 35% in vivo in mouse livers using a lead ABE construct.
The described ABEs and CBEs can target a significant proportion of the adenine and cytosine bases in the human genome. This extensive targetability is demonstrated by the theoretical coverage of their base editors, which significantly exceeds that of traditional SpCas9-based editors.
Metagenomi has discovered and engineered some of the smallest base editors to date, which provides substantial opportunities for vector optimization. These editors, derived from their SMART nuclease platform, are within the size range compatible with AAV delivery methods.
The provided data suggests that Metagenomi's ABE system is capable of efficient editing in primary cells and in vivo, supporting its potential for therapeutic applications. The editing efficiency in mouse liver suggests that a significant proportion of the target cells were successfully edited.
The small size of the SMART base editors, particularly those under 1,000 amino acids, suggests that they could be efficiently packaged into AAV vectors, which are commonly used for in vivo delivery but have size limitations.
Summary: The evidence provided suggests that Metagenomi's base editors have the potential to address some of the limitations of current gene editing technologies, particularly in terms of precision and delivery. The data supporting their claims include in vitro and in vivo studies, showing high editing efficiency and specificity. Their approach could allow for the correction of point mutations associated with a wide range of diseases and the efficient knockdown of genes without the risks associated with double-stranded DNA breaks.
However, while the in vitro and in vivo data seem promising, the true test of these potential advantages will come with further development and clinical trials. The efficiency and specificity of these base editors in humans, their long-term stability and safety, and the ability to scale up for therapeutic applications are all areas that will require additional investigation.
Advantages of Metagenomi's Base Editing Approach
Metagenomi's approach to base editing, as outlined and based on the evidence provided, suggests several potential advantages over established base editing approaches:
Smaller Size for AAV Delivery One of the primary advantages of Metagenomi's base editors is their small size, particularly those derived from the SMART nuclease platform. Smaller nucleases can be more easily packaged into AAV vectors, which have strict size limitations. This can enable the delivery of the entire base editing construct within a single AAV vector, as opposed to splitting it across two vectors, which is often necessary with larger SpCas9-based systems.
Higher Targeting Flexibility Metagenomi's base editors are designed to have extensive genome targetability, potentially allowing for the targeting of a broader range of genomic sites. This could provide the ability to correct a wider array of point mutations across the genome compared to traditional base editors.
Reduced Off-Target Effects The approach emphasizes the selection of highly specific nucleases, which may lead to fewer off-target effects. While off-target effects are a concern for all CRISPR-based editing tools, Metagenomi's meticulous screening for specificity could provide an advantage in terms of safety.
Efficiency Without Double-Stranded Breaks By using engineered nickases that induce single-strand breaks instead of double-stranded breaks, Metagenomi's base editors aim to maintain high editing efficiency while reducing the risks associated with double-stranded DNA breaks, such as translocations or large deletions.
Potential for Multiplexing The presented data suggests that their base editors could be used for multiplexing in primary human T cells with no impact on cell viability. This could be particularly advantageous for therapeutic applications where multiple genes need to be edited simultaneously.
Rapid Optimization Metagenomi's modular, chimeric nuclease platform allows for the rapid optimization of base editors capable of editing various target sites. This can speed up the development of new therapeutic applications.
Novel Deaminases The company has mined a vast library of over three million deaminases, allowing for the identification of novel enzymes that may require minimal engineering to achieve high editing efficiencies.
Versatility for Therapeutic Development The versatility of the platform, as shown by effective editing in primary cells and in vivo, indicates potential use in broad therapeutic applications, including both ex vivo cell therapies and direct in vivo treatments.
While these advantages are promising, it is important to recognize that moving from preclinical success to clinical efficacy and safety is a significant hurdle. The actual therapeutic benefit of these base editors will need to be demonstrated in clinical trials, and issues such as long-term safety, delivery to the correct tissues, and avoidance of immune responses will be critical factors to consider.
Metagenomi's RIGS
Metagenomi's approach to developing RNA-mediated Integration Systems (RIGS), which encompasses both prime editing (Little RIGS) and systems for large genomic integrations (Big RIGS), is detailed and innovative. Here’s how their approach works and how the evidence supports their strategy:
-
Prime Editing (Little RIGS)
Prime editing is a refined method of gene editing that allows for precise edits without the introduction of double-stranded breaks. Metagenomi has created a suite of novel reverse transcriptases (RTs) to pair with their collection of nickases for prime editing. These RTs have been selected for their superior processivity and fidelity, crucial properties for copying the prime editing guide RNA (pegRNA) template into the target DNA without errors. -
Large Genomic Integrations (Big RIGS)
For applications requiring large DNA insertions, such as the addition of an entire gene, Metagenomi is developing Big RIGS. These systems aim to overcome current limitations on the size of the template that can be integrated. By engineering RTs with high processivity and fidelity, and possibly by separating the gRNA and repair template into distinct RNA molecules, Metagenomi is working to enable large-scale genomic integrations. -
Metagenomics Discovery Platform
Metagenomi’s metagenomics platform has been used to mine five million RT candidates to find suitable enzymes for both Little and Big RIGS applications. This vast resource allows for the identification of RTs with the desired biochemical characteristics out of a diverse pool of natural enzymes. -
Programmable Nucleases and Nickases
Their platform of programmable nucleases, which can be converted into nickases, is highly targetable and flexible. This enables the targeting of virtually any genomic site of interest. The use of ultra-small SMART nucleases could further enhance the delivery and specificity of these systems. -
Benchmarking and Optimization
Little RIGS are being benchmarked against industry-standard prime editing systems to identify top-performing systems. The Big RIGS, which are pioneering in the field, are undergoing indication-specific optimization. The goal is to create systems that can enact any type of genomic edit required for therapeutic applications.
The approach and evidence suggest that Metagenomi's RIGS could potentially provide several advantages over existing gene editing methods:
- Versatility: Their RIGS can encode a wide range of genomic modifications, offering a tool to correct nearly any genetic mutation.
- Precision: The high fidelity and processivity of their RTs suggest that these systems can make precise edits with minimal errors.
- Delivery: Their strategy includes developing all-RNA formats for the editing systems, which may simplify the delivery process, particularly for targeting the liver.
- Size of Edits: They are working on overcoming the current size limitations of RNA templates for genomic integrations, which could be a significant advancement for treating diseases caused by large deletions or the need for inserting entire genes.
- Safety: By avoiding double-stranded breaks, these systems may reduce the risks associated with traditional CRISPR/Cas9 editing, such as off-target effects or chromosomal rearrangements.
While these potential advantages are promising, the ultimate test will be the clinical application of these systems. The translation of benchtop innovations to bedside treatments will require thorough validation in clinical trials, ensuring that the edits are safe, stable, and efficacious in the long term. Additionally, while the approach may simplify some aspects of delivery and increase the range of edits, challenges such as immune responses, in vivo efficiency, and tissue-specific targeting still need to be addressed.
Metagenomi's approach to RNA-mediated Integration Systems (RIGS), including Little RIGS for prime editing and Big RIGS for large genomic integrations, could offer several potential advantages over other approaches:
-
Versatility of Edit Types
RIGS can potentially encode a wide array of genomic modifications within an RNA template, providing the flexibility to correct various types of mutations — from point mutations to large insertions and deletions. This goes beyond the capabilities of earlier genome editing technologies that are typically optimized for specific types of edits. -
Avoidance of Double-Stranded Breaks (DSBs)
By using RTs and nickases to incorporate new DNA sequences into the genome, RIGS avoid introducing DSBs, which are the source of many off-target effects and unintended genomic rearrangements associated with CRISPR/Cas9 editing. -
Delivery as RNA
The potential for delivering the entire system as RNA, including the nuclease components, could simplify the delivery process and avoid the integration of foreign DNA into the host genome, which could mitigate some safety concerns. -
Processivity and Fidelity of RTs
Metagenomi's approach to developing RTs with high processivity and fidelity could enable the accurate and efficient copying of longer RNA templates into DNA, necessary for correcting larger genomic segments and potentially improving the efficacy of the edits. -
Large Genomic Integrations
The Big RIGS technology aims to enable the integration of large DNA segments into the genome, which could facilitate the treatment of diseases that require the addition of complete genes, a challenge for current gene therapy approaches that rely on homology-directed repair mechanisms. -
Targeting Precision with Programmable Nucleases/Nickases
Utilizing their proprietary nucleases, which can be programmed to target specific genomic sites, increases the precision of the edits and reduces the likelihood of off-target effects compared to less specific genome editing tools. -
Potential for Multiplexing
The RIGS platform may enable the simultaneous introduction of multiple edits, which is beneficial for complex genetic diseases that require multiple genomic modifications for treatment. -
Packaging Efficiency for Delivery Vectors
For Little RIGS, the use of ultra-small RTs could lead to more efficient packaging within viral vectors such as AAVs, which have packaging size limitations.
The potential advantages outlined by Metagenomi's approach align with the ongoing evolution of gene editing technologies towards higher precision, safety, and versatility. However, the transition from promising preclinical results to clinical success is complex, and these technologies will need to demonstrate safety, efficacy, and deliverability in a clinical setting. The potential for off-target effects, immune responses, and challenges associated with in vivo delivery and long-term expression stability will be crucial factors to consider as Metagenomi's RIGS technology progresses toward therapeutic applications.
Metagenomi's Systems for Large Genomic Integration
Metagenomi's approach to developing systems for large genomic integration, such as CRISPR-associated transposases (CASTs), appears to focus on overcoming limitations of current gene therapy methods, especially for diseases that require the introduction of large segments of DNA. Here’s a summary of their strategy:
-
DNA-Templated Integration
Unlike RNA-templated systems that require a reverse transcription step, DNA-templated systems like CASTs can directly integrate large DNA sequences into the genome. This can allow for the incorporation of much larger templates and may circumvent some of the limitations associated with RNA-mediated systems, such as the size of the template that can be effectively copied and integrated. -
Targeting Complex Genetic Diseases
Metagenomi's CASTs aim to address genetic diseases characterized by a variety of mutations, such as cystic fibrosis, where multiple specific edits might be impractical. Introducing a healthy copy of a gene could offer a universal therapeutic strategy regardless of the mutation present. -
Programmable Integration
CASTs offer programmable and site-specific integration, which is an exciting development compared to non-specific integrations by older transposase or lentiviral systems that have previously resulted in adverse events. This could lead to safer gene therapies by targeting "safe harbor" loci in the genome, reducing the risk of insertional mutagenesis. -
Development of Novel Systems
By mining their metagenomic library, Metagenomi has identified natural variants of transposases that can be engineered for activity in human cells. This approach may allow for the integration of large transgenes with higher accuracy and efficiency. -
Combination Approaches
Metagenomi is also exploring the use of recombinases and other mobile elements in combination with programmable nucleases to achieve large genomic integrations. This could enhance versatility and provide solutions where CASTs alone may not be the most suitable option. -
Biotechnological Applications
The development of CASTs and other large integration systems could extend beyond therapeutic applications to areas such as synthetic biology, antibody discovery, and animal model development, indicating a broad utility for these technologies.
The evidence presented suggests that Metagenomi’s novel CAST systems have demonstrated the capability for targeted integration of transgenes into human cells, which is a significant step forward in the genome editing field. If these results can be replicated and scaled up, they could indeed provide a new class of genome editing therapeutics capable of addressing complex genetic diseases with large genomic integrations. However, the efficiency, specificity, long-term stability, and safety of these integrations in vivo, as well as the potential immune responses to such interventions, will need to be thoroughly investigated in further preclinical and clinical studies.
Metagenomi's CAST integration systems
Metagenomi's approach to large genomic integrations using CRISPR-associated transposases (CAST) and other DNA-templated systems offers several potential advantages compared to other gene therapy approaches:
-
Site-Specific Integration
Unlike viral vectors, which can integrate randomly and potentially disrupt important genes or regulatory regions, CASTs are designed to integrate DNA at precise locations in the genome. This specificity could minimize the risk of insertional mutagenesis and its associated complications. -
Capacity for Large DNA Sequences
CASTs have the potential to integrate significantly larger DNA sequences than traditional CRISPR systems or other gene therapy methods. This could enable the introduction of entire genes, which is especially useful for treating diseases that result from large deletions or complex genetic rearrangements. -
Reduced Dependence on Host Repair Mechanisms
DNA-templated integration systems like CASTs do not rely on the host cell's DNA repair mechanisms to the same extent as other genome editing approaches, potentially reducing the toxicity and increasing the efficiency of the integration process. -
Universal Therapeutic Strategy
For diseases with many different causative mutations, like cystic fibrosis, integrating a healthy copy of the gene could provide a 'one-size-fits-all' treatment, rather than developing multiple specific therapies for different mutations. -
Avoidance of Double-Stranded Breaks (DSBs)
CASTs can integrate DNA without creating DSBs, which are the source of many of the risks associated with CRISPR/Cas9 editing. This could make CAST-based therapies safer by avoiding the unintended consequences of DSBs. -
Potential for Targeted Gene Addition
By targeting 'safe harbor' sites in the genome, CASTs can potentially add therapeutic genes without disrupting endogenous genes or regulatory elements, improving the safety profile of the treatment. -
Flexibility and Versatility
The approach is flexible, allowing for the development of both RNA and DNA-templated systems, which may have different advantages in various therapeutic contexts. -
Scalability
The DNA-templated approach may be more straightforward to scale up for therapeutic production compared to RNA-based systems, which may require more complex synthesis and stability considerations. -
Broad Applicability
The technology could have applications beyond human therapeutics, including in biotechnology fields such as synthetic biology, functional genomics, and more.
Despite these potential advantages, it is important to note that clinical translation involves many challenges. These include ensuring that the integrations are stable and do not lead to long-term adverse effects, confirming that the integrated genes are expressed at therapeutic levels without disrupting the expression of other important genes, and avoiding immune responses to the integrated sequences or the proteins they encode. Additionally, manufacturing and regulatory hurdles for new genetic medicines can be substantial. As such, while the preclinical evidence is promising, comprehensive clinical studies will be necessary to validate these potential advantages in a therapeutic setting.
Potential for Therapeutic Development
The successful demonstration of targeted integration lays the groundwork for therapeutic-driven optimization. The next steps would likely focus on increasing efficiency, ensuring long-term expression and safety, and possibly developing single vector systems.
The evidence supports the potential advantages of Metagenomi's CAST technology for integrating large DNA cargos with specificity. However, the relatively low integration efficiency highlights the need for further research and optimization to reach the levels required for clinical application. Additionally, this is preclinical proof-of-capability, and translating these findings into safe and effective therapies will involve overcoming challenges related to immune responses, long-term stability, and the control of gene expression levels post-integration. The continued discovery and development of novel genome editing systems through Metagenomi's metagenomics platform suggest a promising trajectory for advancing the field of genome editing.
Weekly analyses of biotech startups, generated by AI
Receive high quality, AI-generated analyses of biotech startups, public companies and scientific papers each week.
Market Overview in Hemophilia A
Hemophilia A is a genetic disorder caused by missing or defective Factor VIII, a clotting protein. The condition manifests in an inability to form blood clots effectively, leading to prolonged bleeding after injury or surgery and spontaneous bleeding, especially into joints and muscles. Severe cases may experience frequent joint bleeds, leading to chronic joint disease and pain. With appropriate treatment, individuals with Hemophilia A can lead relatively normal lives. However, without management, the condition can result in chronic joint damage, life-threatening hemorrhages, and significantly reduced quality of life.
Hemophilia A affects around 1 in 5,000 to 10,000 male births worldwide. An estimated 20,000 individuals in the United States live with Hemophilia A. Globally, the number is estimated at over 400,000.
Symptoms include
- Prolonged bleeding from cuts or injuries
- Spontaneous bleeding without apparent cause
- Large or deep bruises
- Painful, swollen joints due to internal bleeding
- Blood in urine or stool
- Frequent nosebleeds
Standard of care includes:
- Prophylaxis: Regular infusions of recombinant Factor VIII are the current standard to prevent bleeding episodes. Prophylactic treatment to prevent bleeds is particularly important for severe cases.
- On-Demand Treatment: Infusions given to stop bleeding when it occurs.
- Bypassing Agents: Used in cases where inhibitors to Factor VIII are present.
Gene therapies like valoctocogene roxaparvovec aim to introduce a functional FVIII gene to produce the clotting factor endogenously. These therapies have shown promise in maintaining therapeutic levels of FVIII and reducing bleeding events. Durability of expression and immune responses are key areas of ongoing research. Novel therapies exploring RNA-based treatments or CRISPR-based gene editing are also being developed for a more durable and potentially curative option:
- Gene Therapy: Approaches like valoctocogene roxaparvovec aim to introduce a functional FVIII gene into the patient's liver cells for long-term production of the clotting factor.
- Antibody Therapy: Emicizumab, a non-factor therapy, mimics the function of Factor VIII and offers prophylactic treatment without the risk of developing inhibitors.
- New Factor Concentrates: Long-acting recombinant Factor VIII products that require less frequent infusions.
- RNAi Therapeutics: Targeting antithrombin RNA to increase thrombin generation in patients with inhibitors.
- CRISPR/Cas9 Gene Editing: Permanent correction of the genetic defect at the DNA level, still largely in the experimental phase.
While replacement therapy is effective, it is not a cure and requires lifelong treatment. The risk of inhibitor development and the high cost of therapy remain significant challenges, along with the need for effective treatments for children and those with inhibitors. Gene therapy offers potential but faces issues such as potential waning effects, immune responses, and re-treatment limitations. Below are some notable aspects of the market:
- High Treatment Costs: Hemophilia treatment is one of the most expensive in medicine, with costs that can exceed $1 million per patient annually.
- Unmet Needs: Despite advances, there are significant unmet needs for therapies that provide constant Factor VIII levels and are suitable for patients with inhibitors.
- Growing Gene Therapy Space: Gene therapy is a rapidly advancing field, with several candidates in late-stage clinical trials promising potential long-term solutions.
- Market Growth: The hemophilia treatment market is expected to grow, driven by the introduction of gene therapies and the need for novel treatment options.
- Regulatory Environment: Regulatory bodies have shown a willingness to fast-track promising therapies for Hemophilia A due to its severe nature and high treatment burden.
The Hemophilia A market is at a transformative stage, with potential shifts from conventional therapies to longer-acting and potentially curative treatments such as gene therapies and gene editing. These advances offer the hope of reducing the treatment burden and improving the quality of life for patients with Hemophilia A. However, these therapies come with high costs and complex manufacturing processes, which may pose challenges for widespread adoption. The successful commercialization of these products will depend on their long-term efficacy, safety profiles, and cost-effectiveness compared to existing treatments.
Gene Therapies for Hemophilia A
Gene therapies for hemophilia A aim to provide a long-term solution to the underlying cause of the disease: the lack of functional Factor VIII (FVIII) protein. By introducing a functional FVIII gene into the patient's own cells, the body can potentially produce its own FVIII, mitigating or even eliminating the need for regular factor infusions. Here are several gene therapy approaches that have been in development:
-
AAV-Mediated Gene Therapies
- Valoctocogene Roxaparvovec (Roctavian) by BioMarin Pharmaceutical: Uses an Adeno-Associated Virus (AAV) vector to deliver a B-domain-deleted version of the human FVIII gene to the patient's liver cells. Received conditional approval in Europe and the United States. Noted decline in FVIII levels over time in some patients.
- Etranacogene Dezaparvovec (Hemgenix) by CSL Behring/uniQure: Uses an AAV vector to deliver a codon-optimized FVIII transgene. Has demonstrated potential for sustained therapeutic expression of FVIII with a good safety profile in clinical trials.
- SPK-8011 by Spark Therapeutics: Investigational AAV-mediated gene therapy delivering the FVIII gene. Has shown potential in early-phase clinical trials to increase FVIII activity levels with a reasonable safety profile.
-
Non-AAV Gene Therapies
- FLT180a by Freeline Therapeutics: AAV-mediated gene therapy using a different capsid and promoter to drive FVIII expression. Aims to achieve normal levels of FVIII activity with a single treatment.
-
Gene Editing Therapies
- CRISPR/Cas9-based Therapies: Rather than delivering a copy of the FVIII gene, aims to correct the gene directly within the patient's cells. Could offer a one-time treatment that permanently corrects the genetic defect.
-
Challenges and Considerations
- Durability of expression and immunogenicity are major concerns.
- Liver toxicity and high costs are also notable challenges.
-
Promising Aspects
- Potential for one-time treatment and improvement in quality of life.
- Could treat a broader range of mutations compared to targeted therapies.
The gene therapy market for hemophilia A is poised to grow as these therapies progress through clinical development and receive regulatory approvals. Their success will depend on overcoming challenges related to durability of expression, immunogenicity, and accessibility. Each new therapy will need to demonstrate a favorable risk-benefit profile to gain acceptance.
There are several safety concerns associated with gene therapies for hemophilia A that researchers, clinicians, and regulatory agencies monitor closely:
- Immunogenicity: Potential immune response to the AAV vector or the newly produced FVIII protein, limiting efficacy and posing safety risks.
- Liver Toxicity: Liver enzyme elevation indicative of hepatotoxicity can occur, likely due to immune response against AAV-transduced hepatocytes.
- Insertional Mutagenesis: Small risk of AAV vector integration into the host genome, potentially disrupting gene function and leading to oncogenesis.
- Germline Transmission: Theoretical risk of gene therapy vectors being incorporated into sperm or eggs, leading to transmission of the inserted gene to offspring.
- Vector Shedding: Possibility of shedding gene therapy vectors through bodily fluids, potentially exposing others to the vector.
- Development of Inhibitors: Risk of patients developing inhibitors against the therapeutic FVIII protein, especially in cases of overexpression.
- Durability of Expression: Uncertainty about how long the therapeutic gene will continue expressing FVIII protein at therapeutic levels.
- Risk of Thrombosis: Overexpression of FVIII can potentially lead to thrombotic events.
- Manufacturing Consistency: Variability in gene therapy products due to complex manufacturing processes.
- Pre-existing Immunity: Effectiveness of treatment can be reduced for patients with pre-existing immunity to AAV vectors.
- Efficacy in the Presence of Inhibitors: Patients with inhibitors to FVIII may not benefit from gene therapies that restore endogenous FVIII production.
Clinical trials for gene therapies in hemophilia A are designed to carefully monitor patients for these and other potential safety issues. Long-term follow-up studies are also a key part of understanding and managing the risks associated with gene therapies.
Metagenomi's Hemophilia A approach
-
Gene Editing Approach
- Permanent Integration: Metagenomi’s approach involves inserting a functional FVIII gene into a safe harbor location within the albumin gene in the liver cells. This is expected to provide stable, long-term expression of FVIII, in contrast to the episomal expression from current AAV gene therapies which may diminish over time.
- Endogenous Promoter: By using the albumin promoter, a strong and naturally occurring promoter, the therapy aims to ensure consistent and high-level expression of the FVIII gene.
- Avoiding Immune Response: The strategy is to integrate the FVIII gene into the liver cells' genome, potentially reducing the risk of immune responses that can occur with episomal gene therapies.
- Pediatric Application: The integrated gene is replicated during cell division, which makes this approach potentially suitable for children, unlike AAV therapies that are less effective due to liver growth diluting the episomal genes.
-
Advantages Over Other Modalities
- Long-term Efficacy: The knock-in approach is designed to achieve stable expression levels that could transform severe hemophilia into a milder form or potentially provide a functional cure.
- Reduced Treatment Burden: With stable gene expression, the need for regular infusions or injections is eliminated, significantly reducing the treatment burden.
- Safety: Targeting a safe harbor locus minimizes the risk of insertional mutagenesis and oncogenesis, which can be a concern with random integrations.
- Economic Efficiency: While gene editing is a complex and costly procedure, it may prove to be more cost-effective in the long run compared to lifelong infusions or treatments with bi-specific antibodies.
- Applicability to Various Mutations: Instead of developing different treatments for various FVIII mutations, a single gene editing therapy could potentially treat all patients with Hemophilia A.
-
Evidence Supporting the Approach
- Preclinical Studies: The presented data from mouse and non-human primate (NHP) models show that the FVIII gene can be successfully integrated and expressed at therapeutic levels.
- Safety Profile: The approach has not shown any notable safety concerns in the preclinical studies, with no significant alterations in liver enzymes or albumin levels post-treatment.
- Durability of Expression: The data suggest that the expression of the integrated FVIII gene remains stable over time, which is crucial for the longevity of the therapeutic effect.
Primary Hyperoxaluria Type 1 (PH1)
Primary Hyperoxaluria Type 1 (PH1) is a rare genetic disorder that affects the metabolism, leading to excessive production and accumulation of oxalate. It is caused by a deficiency of the liver enzyme alanine:glyoxylate aminotransferase (AGT), due to mutations in the AGXT gene. This enzyme deficiency results in the overproduction of oxalate, a substance that, when present in high amounts, combines with calcium to form calcium oxalate crystals that can lead to kidney stones and kidney damage.
The prognosis for individuals with PH1 varies depending on the severity of the condition and when treatment is initiated. If left untreated, PH1 can lead to end-stage renal disease (ESRD) usually in childhood or young adulthood. Systemic oxalosis, where oxalate crystals accumulate in other organs, can occur once the kidneys fail, leading to bone disease, anemia, skin ulcers, and heart and eye problems, further complicating the condition.
PH1 is estimated to affect 1 to 3 people per million, with higher prevalence in certain regions due to founder effects or consanguinity. It accounts for a significant proportion of pediatric ESRD cases.
The standard of care includes aggressive hydration to reduce oxalate concentration in urine, use of citrate to prevent stone formation, pyridoxine (Vitamin B6) in responsive patients, and dialysis in cases of ESRD. Liver transplantation has been a treatment option, as the liver is the site of the defective enzyme; dual liver-kidney transplantation may be considered for those with significant kidney damage.
Despite advancements, there remains an unmet medical need for effective treatments that can prevent the overproduction of oxalate and protect kidney function without the need for lifelong treatments or invasive procedures like organ transplantation.
Emerging therapies include gene therapy approaches that aim to correct the underlying genetic defect and prevent the overproduction of oxalate. Lumasiran, an RNA interference (RNAi) therapy, targets the HAO1 gene to decrease the production of glycolate oxidase, thus reducing oxalate production. Nedosiran, another RNAi therapeutic, inhibits lactate dehydrogenase, which is also involved in oxalate production. These therapies aim to reduce the oxalate burden significantly, potentially changing the disease's natural history. However, issues such as long-term efficacy, accessibility, cost, and the need for ongoing treatment persist, highlighting the ongoing need for novel therapeutic approaches.
Therapeutic Rationale for Gene Editing in Primary Hyperoxaluria Type 1
The therapeutic rationale for using gene editing to durably knock down HAO1 in Primary Hyperoxaluria Type 1 (PH1) is rooted in addressing the underlying metabolic defect causing the disease. PH1 is characterized by mutations in the AGXT gene, leading to dysfunctional alanine glyoxylate aminotransferase and subsequent accumulation of glyoxylate. This excess glyoxylate is converted to oxalate, forming insoluble calcium oxalate crystals in the kidneys.
Metagenomi is partnered with Moderna for the PH1 program. A summary of Metagenomi's approach is as follows:
- Targeted Knockdown of HAO1: Aiming to reduce glyoxylate accumulation by knocking down HAO1, which encodes glycolate oxidase (GO), thus preventing its conversion to oxalate.
- Delivery via Lipid Nanoparticles (LNPs): Delivering mRNA for a nuclease with a guide RNA targeting HAO1, encapsulated within LNPs, to hepatocytes.
- Creating Double-Stranded Breaks in HAO1: The nuclease induces double-stranded breaks in HAO1, leading to its silencing through DNA repair mechanisms.
- Durability of Effect: Aiming for permanent reduction in HAO1 expression, offering potential lifelong benefits.
Safety and Efficacy
- Human Genetic Observations: Safety supported by individuals lacking a functional HAO1 gene without adverse effects.
- siRNA Clinical Data: Existing data support the safety of HAO1-targeting therapies.
- Efficient and Specific Editing: Demonstrated efficient, dose-dependent, and specific editing of HAO1.
- Preclinical Proof-of-Concept: Efficacy shown in reducing urinary oxalate levels in AGXT knockout mice, a PH1 model.
Advantages Over Current Treatments
- Permanent Solution: Offers a potential one-time, permanent solution compared to ongoing treatments.
- Potential for Broader Efficacy: Could address a wider range of PH1 mutations.
- Reduced Treatment Burden: A single administration reduces the treatment burden and improves quality of life.
Overall, the gene editing approach to knock down HAO1 in PH1 represents a novel and potentially transformative therapy. It targets the metabolic basis of the disease with the prospect of a durable, once-off treatment, offering significant advantages over current management strategies.
Risks in Gene Editing Approach for PH1
The gene editing approach to durably knock down HAO1 in Primary Hyperoxaluria Type 1 (PH1) is promising, but it comes with potential scientific and clinical risks crucial for evaluating the treatment's safety and efficacy.
- Off-Target Effects: Risk of unintended genomic sites being impacted, potentially leading to unforeseen genetic alterations.
- Immune Responses: Potential for immune responses due to the introduction of foreign mRNA and guide RNA, despite the general safety of LNPs.
- Durability and Efficiency of Knockdown: Need to evaluate the long-term persistence and efficiency of HAO1 knockdown in humans.
- Unintended Consequences of HAO1 Knockdown: Unknown long-term systemic effects of HAO1 knockdown in a broader patient population.
- Impact on Liver Regeneration and Function: Further investigation required on the impact of editing on liver function over time.
- Risk in Specific Populations: Safety and efficacy in specific populations like children, the elderly, or those with comorbid conditions need careful evaluation.
- Long-term Safety Data: Essential to understand potential late-onset effects or complications.
- Regulatory and Ethical Considerations: Necessity to ensure compliance with regulatory guidelines and address ethical considerations.
- Technical Challenges in Delivery and Administration: Critical to have effective and targeted delivery to the appropriate cells.
Transthyretin Amyloidosis (ATTR)
Transthyretin Amyloidosis (ATTR) is a progressive and often fatal disease characterized by the accumulation of amyloid fibrils, primarily composed of transthyretin (TTR) protein, in various tissues and organs. It significantly impacts clinical outcomes, particularly affecting the heart and peripheral nerves. ATTR leads to progressive organ dysfunction, affecting cardiac and neurological functions, with a life expectancy of typically 3-5 years after diagnosis for cardiac manifestations.
There are two primary types of ATTR:
- ATTRv (Variant ATTR): Caused by genetic mutations in the TTR gene, leading to unstable TTR proteins that misfold and aggregate.
- ATTRwt (Wild-Type ATTR): Associated with aging, where non-mutated TTR proteins misfold and accumulate, especially in the heart.
Approximately 50,000 patients globally with ATTRv and 300,000-500,000 with ATTRwt, more common in older adults and certain populations.
Standard of care includes:
- Tafamidis: Stabilizes TTR tetramers to prevent aggregation. Approved for ATTR cardiomyopathy.
- Antisense Oligonucleotides and siRNA Therapies: Such as inotersen and patisiran, designed to reduce TTR production. Approved for various ATTR manifestations.
- Supportive Care: Management of symptoms and complications, including heart failure treatment.
Despite treatments, ATTR remains progressive with significant morbidity and mortality. Existing therapies require lifelong treatment with limited efficacy, especially in advanced stages. Challenges include treatment accessibility, need for earlier diagnosis, and impact on cardiac and neurological functions.
Gene Editing in Transthyretin Amyloidosis
The therapeutic rationale for using a gene editing approach to knock down the expression of the TTR gene in Transthyretin Amyloidosis (ATTR) focuses on addressing the root cause of the disease – the misfolding and aggregation of transthyretin protein. Metagenomi is partnered with Ionis in ATTR.
In ATTR, either variant (ATTRv) or wild-type (ATTRwt) TTR proteins misfold and aggregate, forming amyloid deposits in tissues, particularly in the heart and peripheral nerves. The TTR protein is synthesized primarily in the liver.
Current treatments, like tafamidis and similar therapies stabilize the TTR tetramer but do not stop its production. Antisense oligonucleotides and siRNA therapies aim to reduce TTR levels but require ongoing treatment and may not completely halt disease progression.
The benefits to a gene editing approach include:
- Editing the TTR gene in the liver aims to reduce or eliminate the source of misfolded proteins.
- Potentially offers a permanent solution with long-lasting effects, unlike therapies requiring regular administration.
- Lowering TTR levels could prevent or reduce amyloid fibril formation.
- Beneficial for both ATTRv and ATTRwt patients.
While gene editing is a promising, potentially longer-lasting therapy, it comes with additional risks:
- Off-Target Effects: Risk of unintended edits in the genome leading to adverse effects.
- Immunogenicity: Potential immune responses to foreign nucleases.
- Durability of Effect: Uncertainty about long-term stability of the edits.
- Delivery Challenges: Need for efficient targeting and uptake by hepatocytes.
- Unintended Consequences: Potential impact on physiological roles of TTR.
- Regulatory and Ethical Considerations: Strict scrutiny and ethical considerations in gene editing.
The gene editing approach for TTR knockdown in ATTR is innovative and potentially durable, but it carries risks and challenges, particularly in safety and delivery. Compared to existing therapies, it offers sustained benefits but lacks established safety profiles and reversible treatment options. Careful clinical development and safety monitoring are vital for mitigating these risks.
Key license agreements
Moderna Agreement
- Effective from October 29, 2021, this agreement involves collaboration in the research, development, and commercialization of in vivo genome editing therapies.
-
Collaboration Scope
- Exclusive Access: Moderna has exclusive access to the company's technology platform for in vivo gene editing in specific fields.
- Joint Committees: Formation of joint steering, research, and patent subcommittees.
-
Licensing and Ownership
- Non-exclusive Licenses: Granted for background technology and related intellectual property.
- Ownership of Intellectual Property: Specified ownership for Metagenomi and Moderna Program Technology, and joint ownership for certain discoveries.
-
Research and Development
- Collaboration Programs: Involving specific research fields and terms.
-
Financial Terms
- Upfront payments, research cost reimbursements, and convertible promissory note details.
- Milestones and Royalties: Specified fees, milestone payments, and royalties on net sales.
-
Commercialization
- DT Co-Co Program: Co-development and commercialization terms.
- Commercialization Rights: Distribution of rights between the U.S. and other countries.
-
Term and Termination
- Royalty Term: Defined duration post-first commercial sale.
- Termination Rights: Conditions under which either party can terminate the agreement.
-
Key Legal Provisions
- Termination Consequences: Outlines the aftermath of termination.
- Adjustments: Conditions under which Moderna can propose profit-share adjustments.
Affini-T agreement
- The agreement between the company and Affini-T, effective June 14, 2022, focuses on developing gene-edited TCR-based therapeutic products for cancer treatment.
-
Collaboration and Licensing
- Field of Collaboration: Development and commercialization of gene-edited TCR-based therapies for human cancer treatment.
- Joint Steering Committee: Established to oversee collaboration activities.
- License Grants: Options for up to six targets with exclusive or non-exclusive licensing rights for TCR-based therapies.
-
Financial Terms
- Upfront Equity Consideration: 719,920 shares of Affini-T’s common stock valued at approximately $1.3 million.
- Expense Reimbursement: Affini-T to reimburse research expenses, totaling $3.2 million as of September 30, 2023.
- Milestone Payments and Royalties: Includes equity milestone, developmental milestones, regulatory approval milestones, sales-based milestones, and royalties.
-
Research and Development
- Exclusive Option Rights: Affini-T can exercise options for licenses for each target.
- Development and Commercialization: Affini-T is responsible for all development and commercialization efforts.
- Non-compete Provision: Restrictions on exploiting gene-edited products directed to certain targets.
-
Agreement Term and Termination
- Initial Term: Five years from the effective date, with the possibility of extension.
- Termination Rights: Conditions under which either party can terminate the agreement.
- Continuation Option in Case of Breach: Affini-T may continue the agreement at reduced payments upon material breach by the company.
-
Key Provisions
- Royalty Term: Duration of royalty obligations on a target-by-target basis.
- Rights Post-Termination: Specific rights and obligations following termination or expiration of the agreement.
Ionis Agreement
- Effective November 10, 2022, this agreement focuses on developing gene editing therapies.
-
Collaboration and Licensing
- License Grant: Worldwide exclusive, royalty-bearing license granted to Ionis for in vivo gene editing in humans.
- Co-Development and Commercialization: Exclusive option for the company to co-develop and co-commercialize certain products.
- Joint Steering Committee: Established to oversee research and drug discovery activities.
- Target Selection: Up to eight targets with responsibility for selection by Ionis.
-
Financial Terms
- Upfront Payment: $80.0 million received from Ionis.
- Reimbursement for Research Activities: Up to $10.0 million, payable in quarterly installments.
- Milestone Payments: Specified development, regulatory, and sales-based milestones.
- Royalties: On annual net sales of licensed products, with standard reductions.
- Co-Co Option Exercise Fee: Calculated as 50% of Ionis’ costs, reduced by the company’s costs.
-
Research and Development
- Drug Discovery Programs: Ionis responsible for development and commercialization post-candidate identification.
- Exploratory Research Program: Joint optimization and selection activities.
- Duration: Drug discovery activities with a five-year term, extendable under certain conditions.
-
Rights and Obligations
- Manufacturing: The company to manufacture licensed systems and components for Ionis.
- Ionis IP Option: Option for a non-exclusive license for Ionis’ technology for therapeutic products.
-
Term and Termination
- Term: Continuation until the expiration of all royalty terms for each licensed product.
- Termination Rights: Conditions under which either party can terminate the agreement.
- Post-Termination: Transfer of regulatory approvals to the company and a potential royalty-bearing license back to Ionis.
-
Key Provisions
- Research Collaboration: In-depth collaboration in drug discovery and exploratory research.
- Profit and Cost Sharing: Equal sharing of costs and profits in case of co-commercialization.
Competition
Metagenomi faces competition from various entities based on their technological focus and corporate status. Here is an overview:
-
CRISPR/Cas Nuclease Technology
- Public Companies:
- Caribou Biosciences, Inc.
- Editas Medicine, Inc.
- CRISPR Therapeutics AG
- Intellia Therapeutics, Inc.
- Graphite Bio, Inc.
-
Base Editing Technology
- Public Companies:
- Beam Therapeutics Inc.
- Verve Therapeutics, Inc.
-
Prime Editing Technology
- Public Companies:
- Prime Medicine
-
First-Generation Nuclease-Based Genome Editing (ZFNs, Meganucleases, TALENs)
- Public Companies:
- Sangamo Therapeutics, Inc.
- Precision BioSciences, Inc.
- Cellectis S.A.
- bluebird bio, Inc.
-
Gene Therapy, Oligonucleotides, and CAR-T Therapeutic Approaches
- Competition from various companies (specific company status not provided).
-
Genome and Epigenome Editing Therapies
- Private Companies:
- Arbor Biotech
- Chroma Medicine, Inc.
- Mammoth Biosciences
- Scribe Therapeutics
- Tessera Therapeutics
- Tome Biosciences
- Tune Therapeutics, Inc.
In summary, Metagenomi operates in a highly competitive field with various companies employing different gene editing technologies. These include both public and private companies engaged in innovative research and development across several areas, including CRISPR/Cas9, base editing, prime editing, older genome editing technologies, and gene therapy approaches.